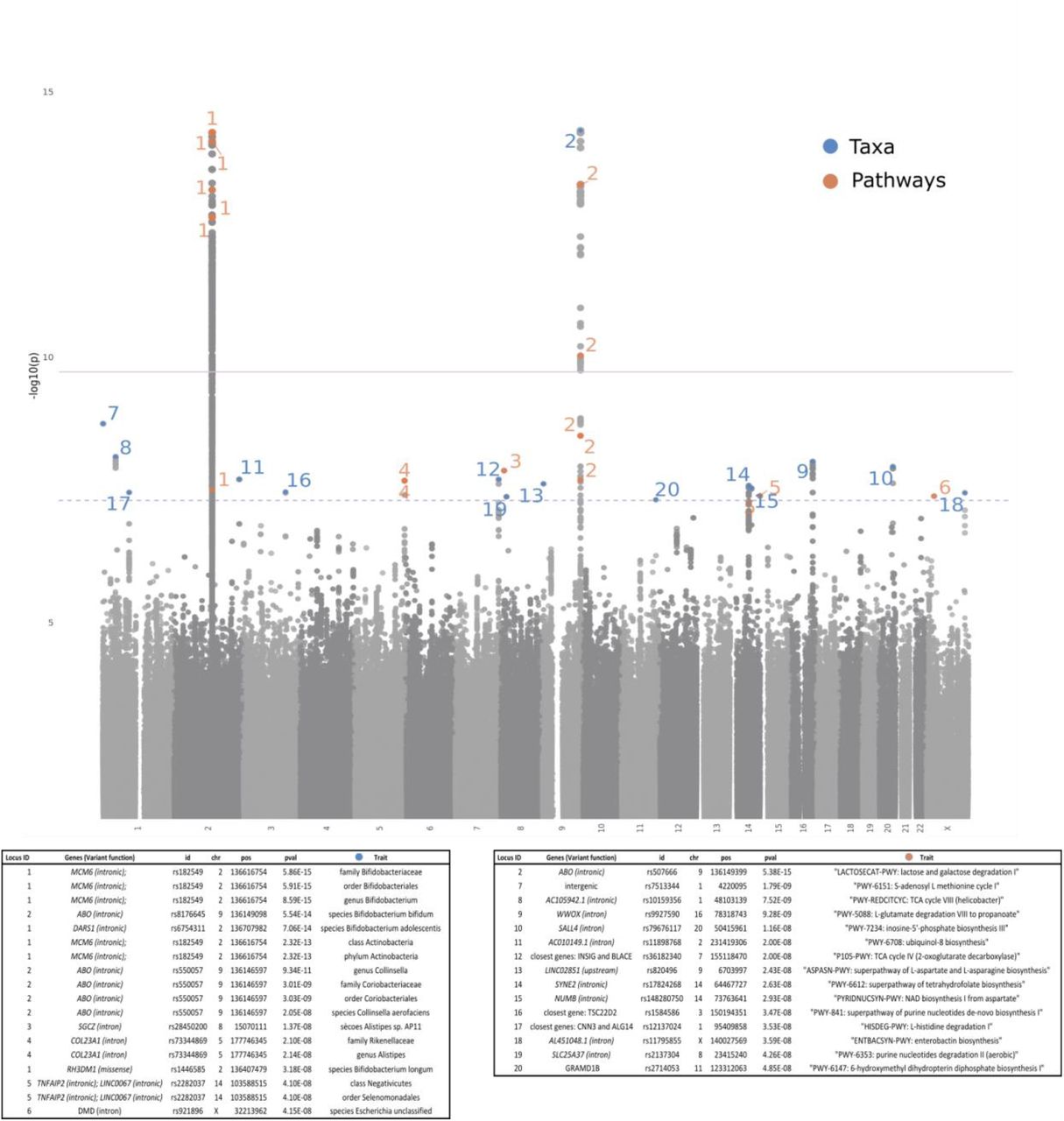
The results of genome-wide association study of effect of host genetics on gut microbiome in the Dutch Microbiome Project cohort are now available on BioRxiv! We studied 207 taxa and 205 pathways from gut microbiomes of 7,738 participants in the Dutch Microbiome Project, a cohort from the Northern Netherlands. We found two obust, study-wide significant (p<1.89×10−10) signals near the LCT and ABO genes were found to affect multiple microbial taxa and pathways, and replicated these results in two independent cohorts.
We found that the LCT locus associations are modulated by host’s lactose intake, while those at ABO reflected participant secretor status determined by FUT2 genotype. Eighteen other loci showed suggestive evidence (p<5×10−8) of association with microbial taxa and pathways. Summary statistics are available on our DMP website.

Comments are closed